A Deep Dive Into the Genetics of Alcohol Consumption
A research group centered at the University of California San Diego School of Medicine has drilled deep into a dataset…
Fast Four Quiz: Precision Medicine in Cancer
Nuisance Seaweed Found to Produce Compounds with Biomedical Potential
A seaweed considered a threat to the healthy growth of coral reefs…
Retired stars join the young stars’ party in the sky: how evolved stars contribute to the early heating of Earth
Researchers from the University of Sheffield and Imperial College London have spotted a ‘retired’ Asymptotic…
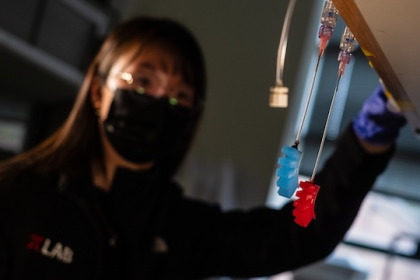
Rice breakthrough could make automated dosing systems universal
Synthetic biologists’ hack blood-glucose reaction to create chemotherapy detector Rice University synthetic biologists have found a way to piggyback on…
Sign Up for Free
Subscribe to our newsletter and don't miss out on our programs, webinars and trainings.
National Chemistry Week reminds us of the significance of chemistry
Chemistry, often hailed as the cornerstone of scientific disciplines, plays a pivotal role in our daily existence, influencing the food…
Your one-stop resource for medical news and education.
Nuisance Seaweed Found to Produce Compounds with Biomedical Potential
A seaweed considered a threat to the healthy growth of coral reefs in Hawaii may possess the ability to produce…
New tools used to identify childhood cancer genes
Using a new computational strategy, researchers at UT Southwestern Medical Center have identified 29 genetic…
New 3D printer uses rays of light to shape objects, transform product design
A new 3D printer uses light to transform gooey liquids into complex solid objects in…
Cotton-Based Hybrid Biofuel Cell Could Power Implantable Medical Devices
A glucose-powered biofuel cell that uses electrodes made from cotton fiber could someday help power…
A Deep Dive Into the Genetics of Alcohol Consumption
A research group centered at the University of California San Diego School of Medicine has…
MIT engineers design flexible “skeletons” for soft, muscle-powered robots
New modular, spring-like devices maximize the work of live muscle fibers so they can be…
This 3D printer can figure out how to print with an unknown material
While 3D printing has exploded in popularity, many of the plastic materials these printers use…
Revolutionary chronic wound treatment could help millions
An effective treatment for chronic wounds that does not involve antibiotics, but an ionised gas…
Millions are at risk using high arsenic water for cooking
The use of water contaminated with higher than recommended levels of arsenic could pose a…
Rice research could advance soft robotics manufacturing, design
Quantitative framework enables tunable elastomer curing via temperature control Te Faye Yap, a Rice University…
Rice breakthrough could make automated dosing systems universal
Synthetic biologists’ hack blood-glucose reaction to create chemotherapy detector Rice University synthetic biologists have found…
BRAIN PROTEIN’S VIRUS-LIKE STRUCTURE MAY HELP EXPLAIN CANCER-INDUCED MEMORY LOSS
In a rare but serious complication of cancer, the body’s own immune system can start…