A fungal origin for coveted lac pigment
The colourful pigment extracted from the lac insect may actually be produced by a symbiotic yeast-like organism living inside the…
Fast Four Quiz: Precision Medicine in Cancer
Bacteria ‘nanowires’ could help scientists develop green electronics
Engineered protein filaments originally produced by bacteria have been modified by scientists…
Nuisance Seaweed Found to Produce Compounds with Biomedical Potential
A seaweed considered a threat to the healthy growth of coral reefs…
An ancient relative of humans shows a surprisingly modern trait
Study finds an archaic hominin had modern dental growth A relative of modern humans that…
Antarctic Ice Core Contains Unrivaled Detail of Past Climate
Donald Voigt, chief scientist at the WAIS Divide Camp, examines an ice core. (Credit: Gifford…
Serious Loneliness Spans the Adult Lifespan but there is a Silver Lining
Feeling alone linked to psychological and physical ills, but wisdom may be a protective factor In recent years, public health…
Sign Up for Free
Subscribe to our newsletter and don't miss out on our programs, webinars and trainings.
Bacteria ‘nanowires’ could help scientists develop green electronics
Engineered protein filaments originally produced by bacteria have been modified by scientists to conduct electricity. In a study published recently…
Rapid Testing for TB Aims to Reduce Drug Resistance, Lower Mortality Rate
Researchers at University of California, San Diego School of Medicine have documented…
Your one-stop resource for medical news and education.
Bacteria ‘nanowires’ could help scientists develop green electronics
Engineered protein filaments originally produced by bacteria have been modified by scientists to conduct electricity. In a study published recently…
Bacteria ‘nanowires’ could help scientists develop green electronics
Engineered protein filaments originally produced by bacteria have been modified by scientists to conduct electricity. In a study published recently…
New tools used to identify childhood cancer genes
Using a new computational strategy, researchers at UT Southwestern Medical Center have identified 29 genetic…
Cotton-Based Hybrid Biofuel Cell Could Power Implantable Medical Devices
A glucose-powered biofuel cell that uses electrodes made from cotton fiber could someday help power…
New 3D printer uses rays of light to shape objects, transform product design
A new 3D printer uses light to transform gooey liquids into complex solid objects in…
A fungal origin for coveted lac pigment
The colourful pigment extracted from the lac insect may actually be produced by a symbiotic…
Combination approach could overcome treatment resistance in deadly breast cancer
QIMR Berghofer-led research in collaboration with Australian oncology company, Kazia Therapeutics, has found that combining…
Tailored brain stimulation treatment results give new hope for people with depression
Medical researchers at QIMR Berghofer have achieved a significant milestone in the treatment of depression,…
Game-Changer in Emergency Medicine: New AI Test Flags Sepsis Hours Before Symptoms Worsen
When the body's reaction to an infection goes awry, it can result in sepsis, a…
Perfumes and lotions disrupt how body protects itself from indoor air pollutants
Fragrances and lotions don't just change the way people smell, they actively alter the indoor…
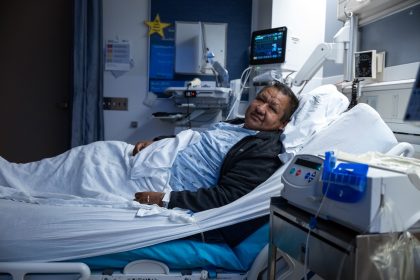
Medical Milestone: Surgeons Perform First-Ever Human Bladder Transplant
In a groundbreaking medical achievement, surgeons at the University of California, Los Angeles (UCLA) Health…
A Downside of Taurine: It Drives Leukemia Growth
A new scientific study identified taurine, which is made naturally in the body and consumed…
How do therapy dogs help domestic abuse survivors receiving support services?
A new exploration of how therapy dogs can create a safe, nonjudgmental environment for survivors…